Selected publications
This only lists our main publications. See Google Scholar for all publications.
2025
-
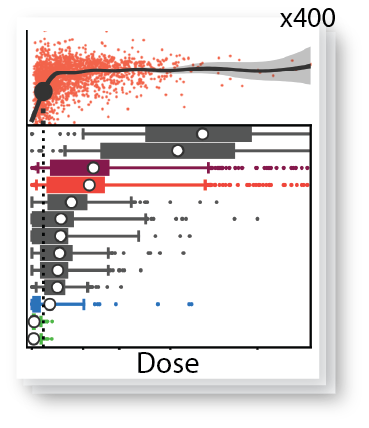 Nature Genetics, Oct 2025
Nature Genetics, Oct 2025A high-throughput technology to study the effect of transcription factor dose. Applied to reprogramming, it reveals how TF dose affects cell fate heterogeneity.
-
 Journal of Open Source Software, Apr 2025
Journal of Open Source Software, Apr 2025A clean and easy way to make complex heatmaps in Python, R and Javascript.
-
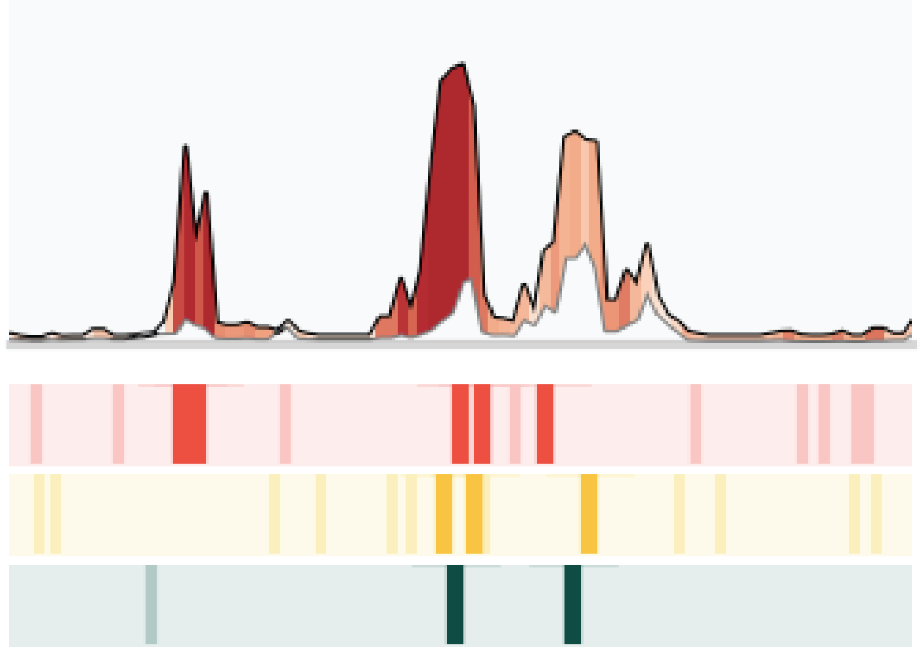 Nature Communications, Jan 2025
Nature Communications, Jan 2025A scale-adaptive machine learning method to link single-cell chromatin accessibility to gene expression. Outperforms peak- and window-based methods by a large margin.
2022
-
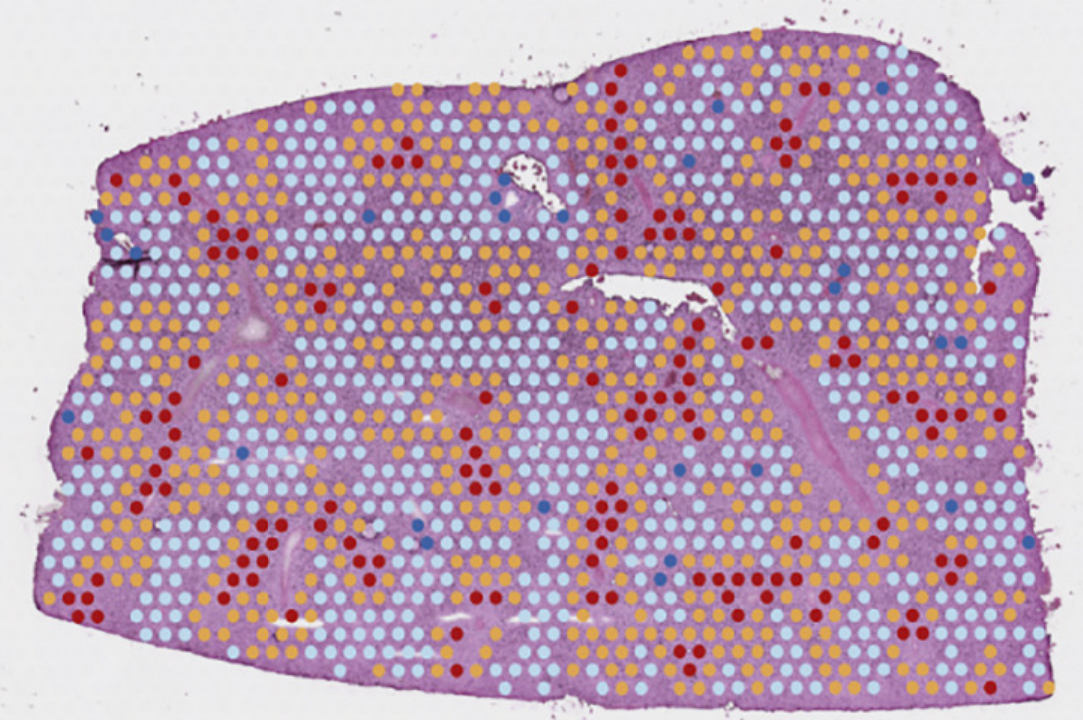 Cell, Jan 2022
Cell, Jan 2022One of the first comprehensive liver cell atlases combining single-cell and spatial transcriptomics with proteomics. Beside being a key resource, it reveals distinct macrophage niches conserved across species.
2021
-
 Nature Communications, Jun 2021
Nature Communications, Jun 2021A flexible simulator for single-cell multi-omics data, useful for benchmarking computational methods. Builds on a detailed model of gene regulation, splicing, and translation.
2020
-
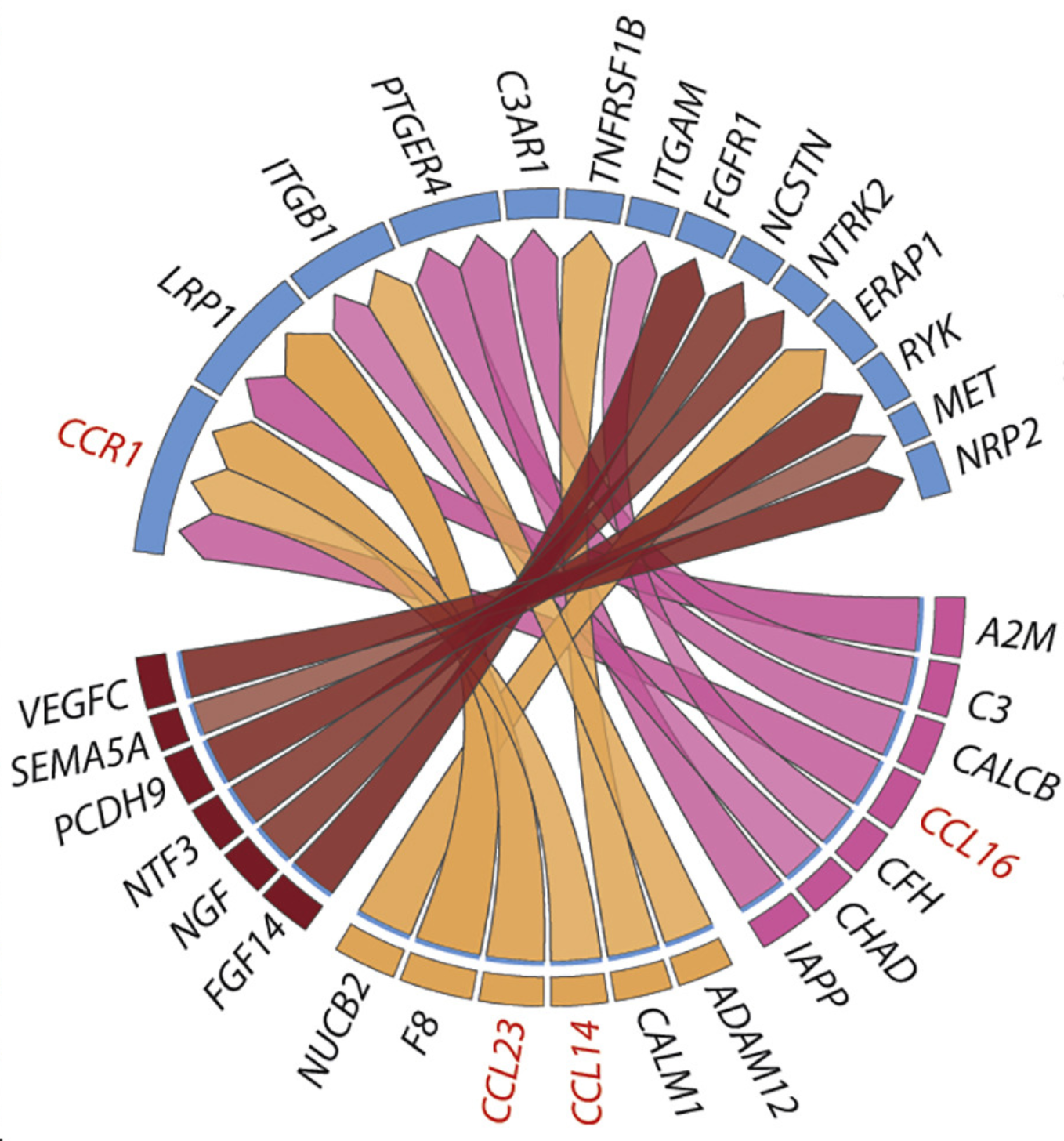 Nature Methods, Feb 2020
Nature Methods, Feb 2020A widely used method to predict cell-cell communication from single-cell data. It uniquely not only looks at ligand-receptor pairs, which is bound to contain false-positives, but also models downstream target gene regulation to ensure the signaling is actively sensed by the cell.
2019
-
 Nature Biotechnology, May 2019
Nature Biotechnology, May 2019The reference benchmark paper for single-cell trajectory inference methods. People love them or hate them, but everyone uses them.
2018
-
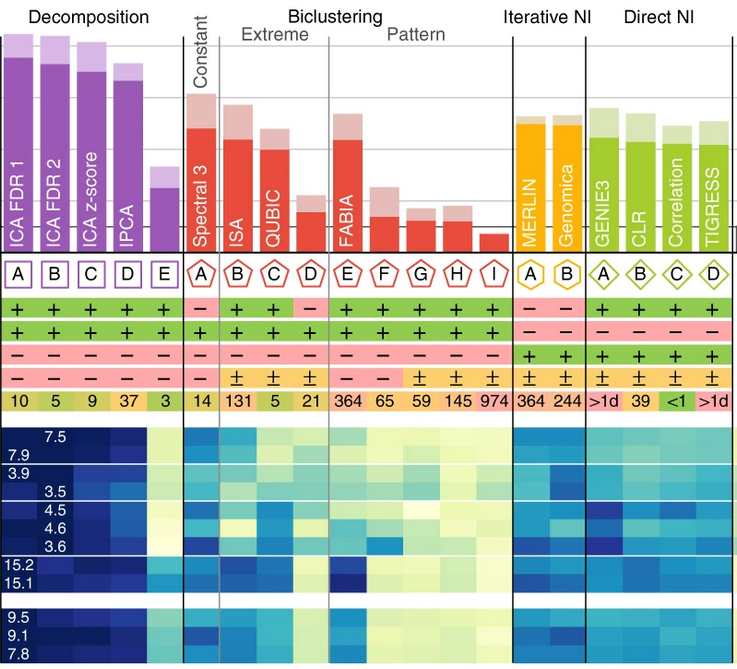 Nature Communications, Mar 2018
Nature Communications, Mar 2018Not all module detection methods are created equal: decomposition methods work best - if you can handle the more complex interpretation.